Tutorial
Welcome to ktplotspy! This is a python library to help visualise CellPhoneDB results, ported from the original ktplots R package (which still has several other visualisation options). Here, we will go through a quick tutorial on how to use the functions in this package.
Import libraries
[1]:
import os
import anndata as ad
import pandas as pd
import ktplotspy as kpy
import matplotlib.pyplot as plt
Prepare input
We will need 3 files to use this package, the h5ad file used for CellPhoneDB,the means.txt, pvalues.txt. deconvoluted.txt is only used for plot_cpdb_chord.
[2]:
os.chdir(os.path.expanduser("~/Documents/Github/ktplotspy"))
# read in the files
# 1) .h5ad file used for performing CellPhoneDB
adata = ad.read_h5ad("data/kidneyimmune.h5ad")
# 2) output from CellPhoneDB
means = pd.read_csv("data/out/means.txt", sep="\t")
pvals = pd.read_csv("data/out/pvalues.txt", sep="\t")
decon = pd.read_csv("data/out/deconvoluted.txt", sep="\t")
Heatmap
The original heatmap plot from CellPhoneDB can be achieved with this reimplemented function.
[3]:
kpy.plot_cpdb_heatmap(pvals=pvals, figsize=(5, 5), title="Sum of significant interactions")
[3]:
<seaborn.matrix.ClusterGrid at 0x17f129150>
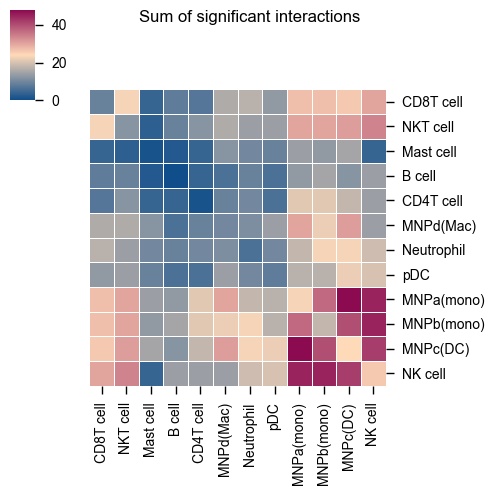
You can also specify specific celltypes to plot.
[4]:
kpy.plot_cpdb_heatmap(
pvals=pvals, cell_types=["NK cell", "pDC", "B cell", "CD8T cell"], figsize=(4, 4), title="Sum of significant interactions"
)
[4]:
<seaborn.matrix.ClusterGrid at 0x17f3b7750>
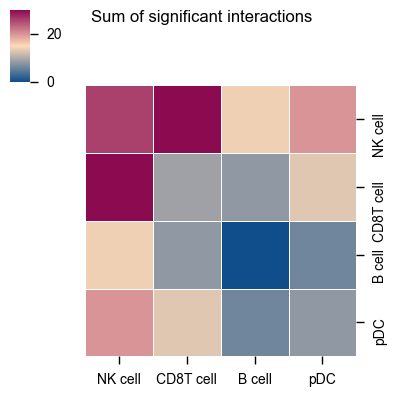
The current heatmap is directional (check count_network and interaction_edges for more details in return_tables = True).
To obtain the heatmap where the interaction counts are not symmetrical, do:
[5]:
kpy.plot_cpdb_heatmap(
pvals=pvals,
figsize=(5, 5),
title="Sum of significant interactions",
symmetrical=False,
)
[5]:
<seaborn.matrix.ClusterGrid at 0x17df3e7d0>
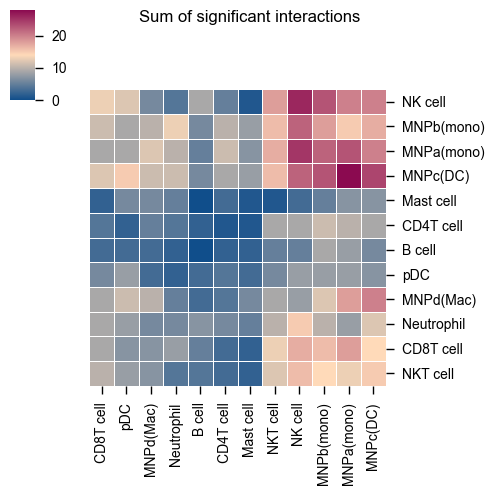
The values for the symmetrical=False mode follow the direction of the L-R direction where it’s always moleculeA:celltypeA -> moleculeB:celltypeB.
Therefore, if you trace on the x-axis for celltype A [MNPa(mono)] to celltype B [CD8T cell] on the y-axis:
A -> B is 18 interactions
Whereas if you trace on the y-axis for celltype A [MNPa(mono)] to celltype B [CD8T cell] on the x-axis:
A -> B is 9 interactions
symmetrical=True mode will return 18+9 = 27
Dot plot
A simple usage of plot_cpdb is like as follows:
[6]:
# TODO: How to specify the default plot resolution??
kpy.plot_cpdb(
adata=adata,
cell_type1="B cell",
cell_type2=".", # this means all cell-types
means=means,
pvals=pvals,
celltype_key="celltype",
genes=["PTPRC", "TNFSF13B"],
figsize=(13, 4),
title="interacting interactions!",
)
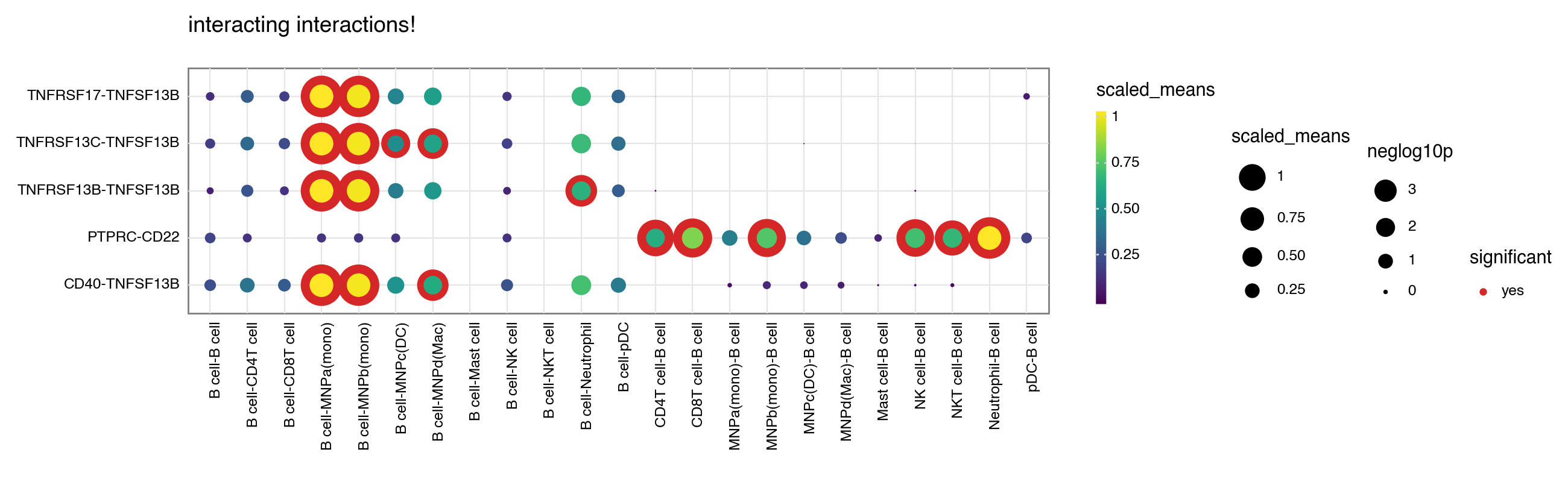
[6]:
<Figure Size: (1300 x 400)>
You can toggle keep_id_cp_interaction to keep the original interaction id. This is useful when there are duplicate interaction names (from cellphonedb V5 onwards).
[7]:
kpy.plot_cpdb(
adata=adata,
cell_type1="B cell",
cell_type2=".", # this means all cell-types
means=means,
pvals=pvals,
celltype_key="celltype",
genes=["PTPRC", "TNFSF13B"],
figsize=(13, 4),
title="interacting interactions!",
keep_id_cp_interaction=True,
)
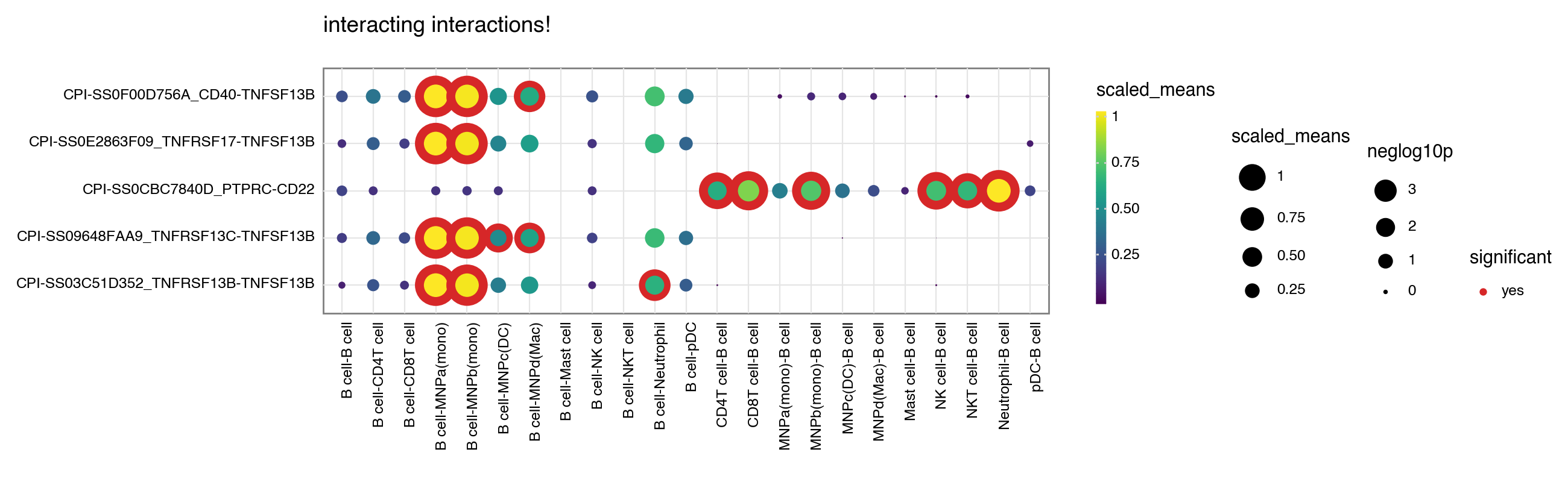
[7]:
<Figure Size: (1300 x 400)>
You can also specify a gene_family.
[ ]:
kpy.plot_cpdb(
adata=adata,
cell_type1=".",
cell_type2=".",
means=means,
pvals=pvals,
celltype_key="celltype",
gene_family="chemokines",
highlight_size=1,
figsize=(20, 8),
)
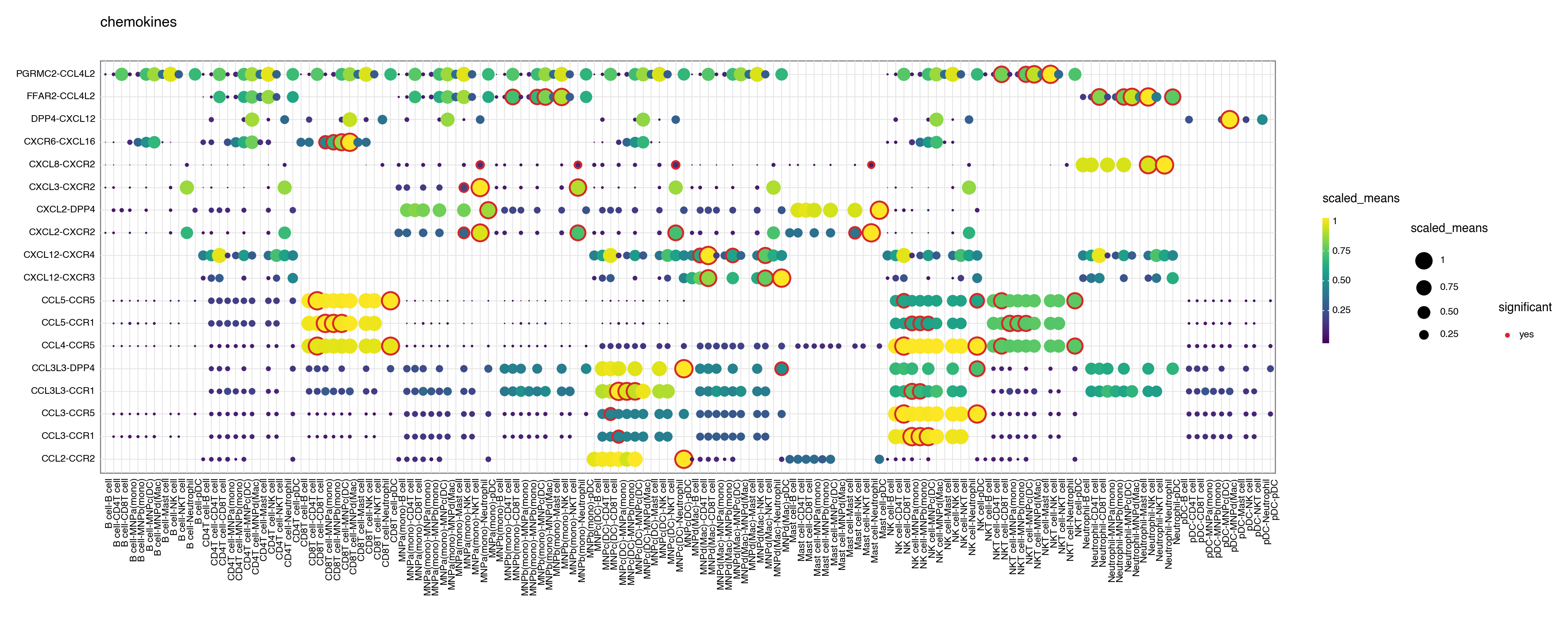
<Figure Size: (2000 x 800)>
<Figure Size: (2000 x 800)>
Or don’t specify either and it will try to plot all significant interactions.
[9]:
kpy.plot_cpdb(
adata=adata,
cell_type1="B cell",
cell_type2="pDC|T",
means=means,
pvals=pvals,
celltype_key="celltype",
highlight_size=1,
figsize=(6.5, 5.5),
)
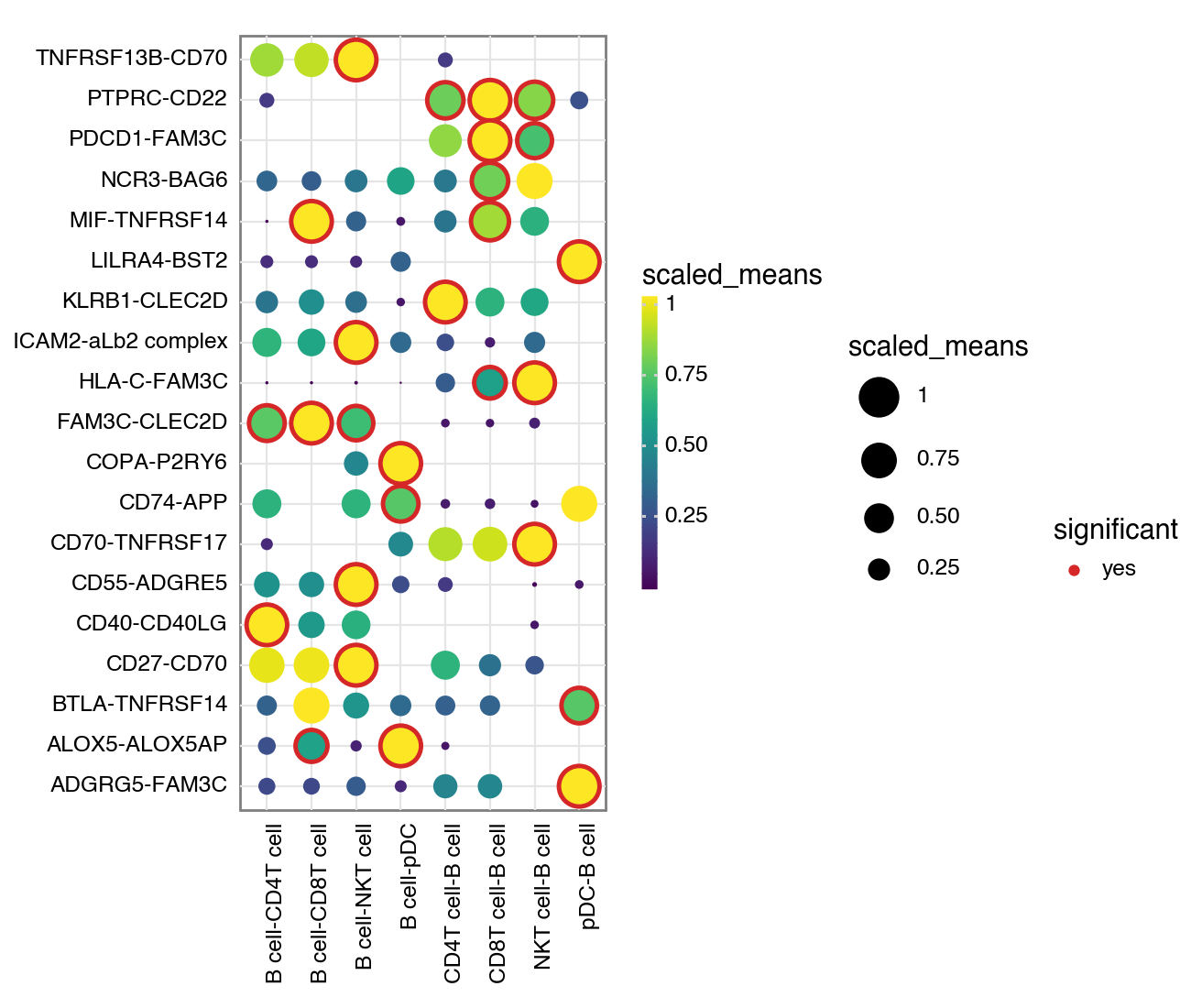
[9]:
<Figure Size: (650 x 550)>
If you prefer, you can also use the squidpy inspired plotting style:
[10]:
kpy.plot_cpdb(
adata=adata,
cell_type1="B cell",
cell_type2=".",
means=means,
pvals=pvals,
celltype_key="celltype",
genes=["PTPRC", "CD40", "CLEC2D"],
default_style=False,
figsize=(13, 4),
)
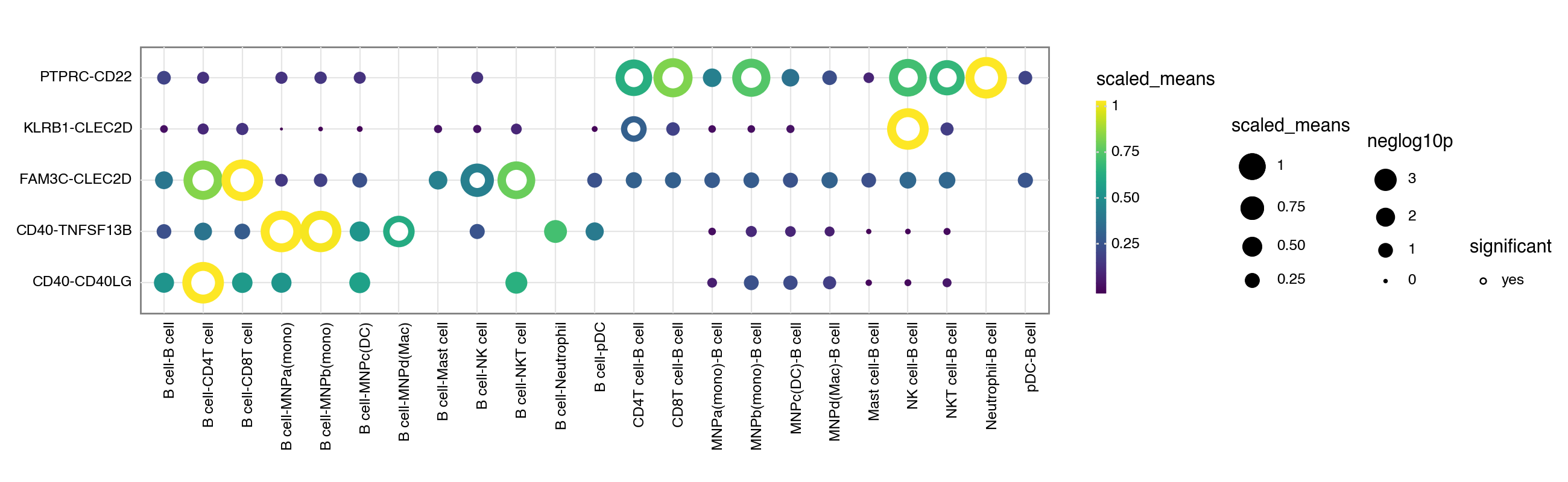
[10]:
<Figure Size: (1300 x 400)>
Chord diagram
There is a preliminary implementation of a chord diagram:
[11]:
kpy.plot_cpdb_chord(
adata=adata,
cell_type1="B cell",
cell_type2=".",
means=means,
pvals=pvals,
deconvoluted=decon,
celltype_key="celltype",
genes=["PTPRC", "CD40", "CLEC2D"],
figsize=(6, 6),
labelposition=50,
)
[11]:
<pycircos.pycircos.Gcircle at 0x28daeea10>
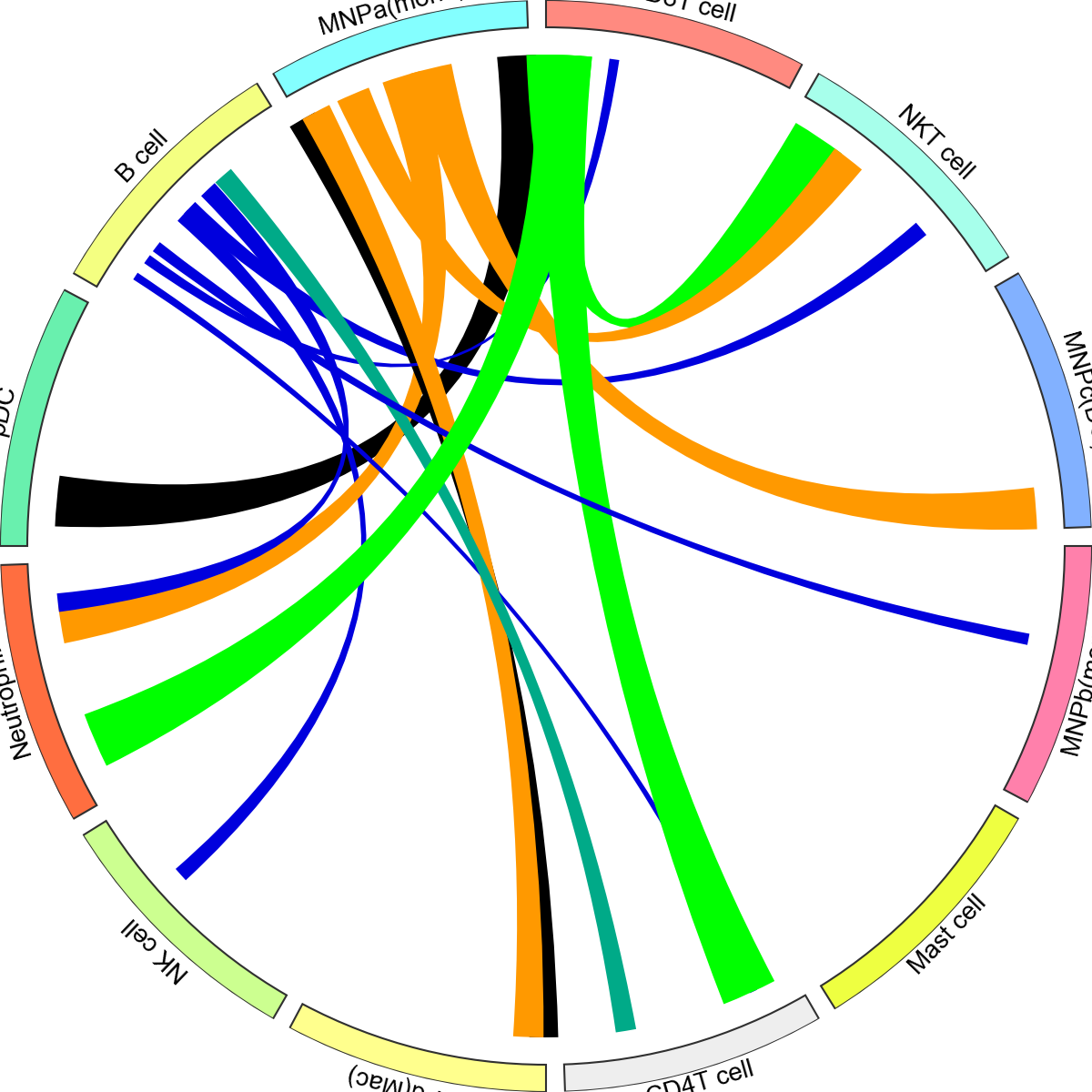
Colour of edges can be changed with a dictionary (below), or with a edge_cmap option:
[12]:
kpy.plot_cpdb_chord(
adata=adata,
cell_type1="B cell",
cell_type2=".",
means=means,
pvals=pvals,
deconvoluted=decon,
celltype_key="celltype",
genes=["PTPRC", "CD40", "CLEC2D"],
edge_cmap=plt.cm.coolwarm,
figsize=(6, 6),
labelposition=50,
)
[12]:
<pycircos.pycircos.Gcircle at 0x28dbeabd0>
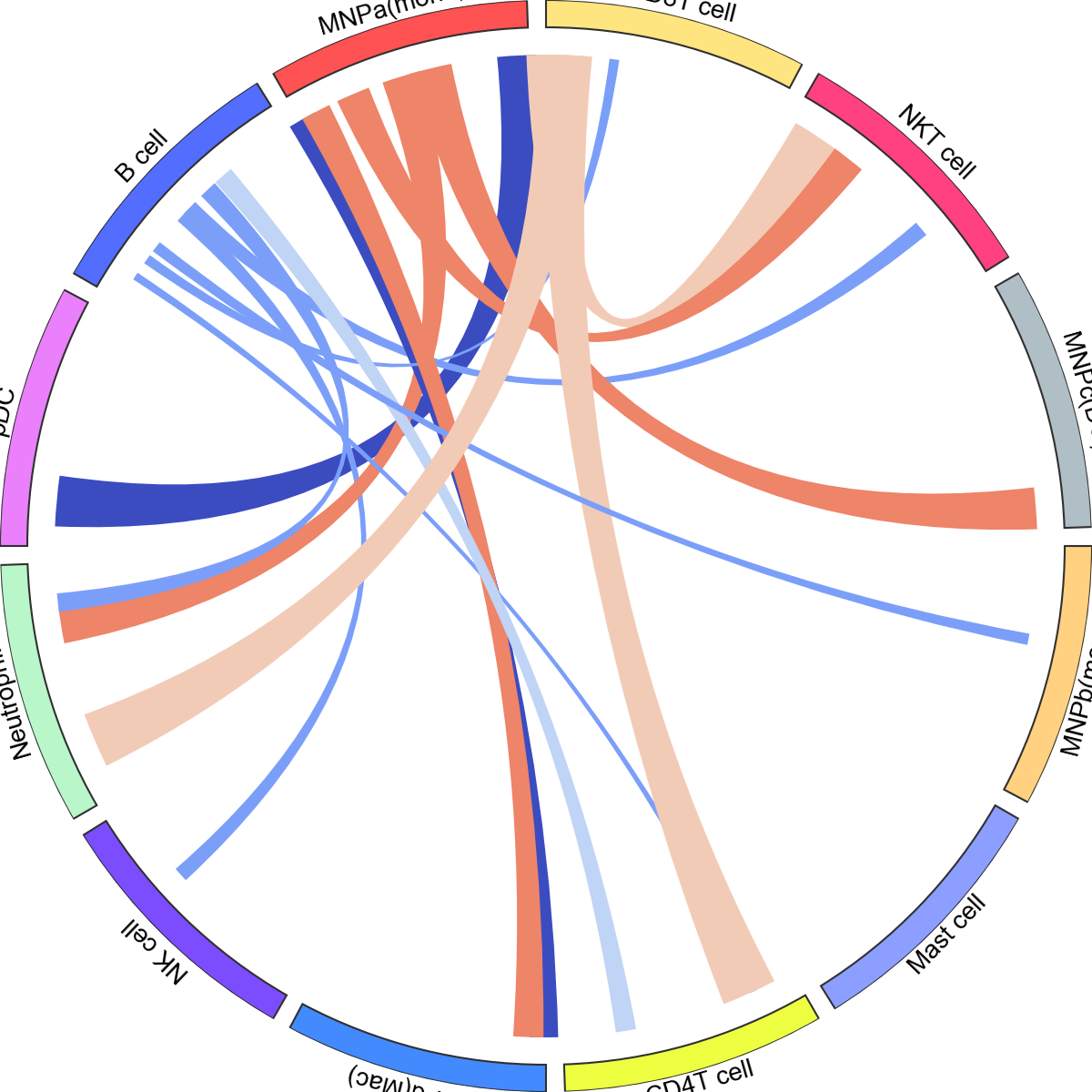
If your adata already has e.g. adata.uns['celltype_colors'], it will retrieve the face_colours correctly:
[13]:
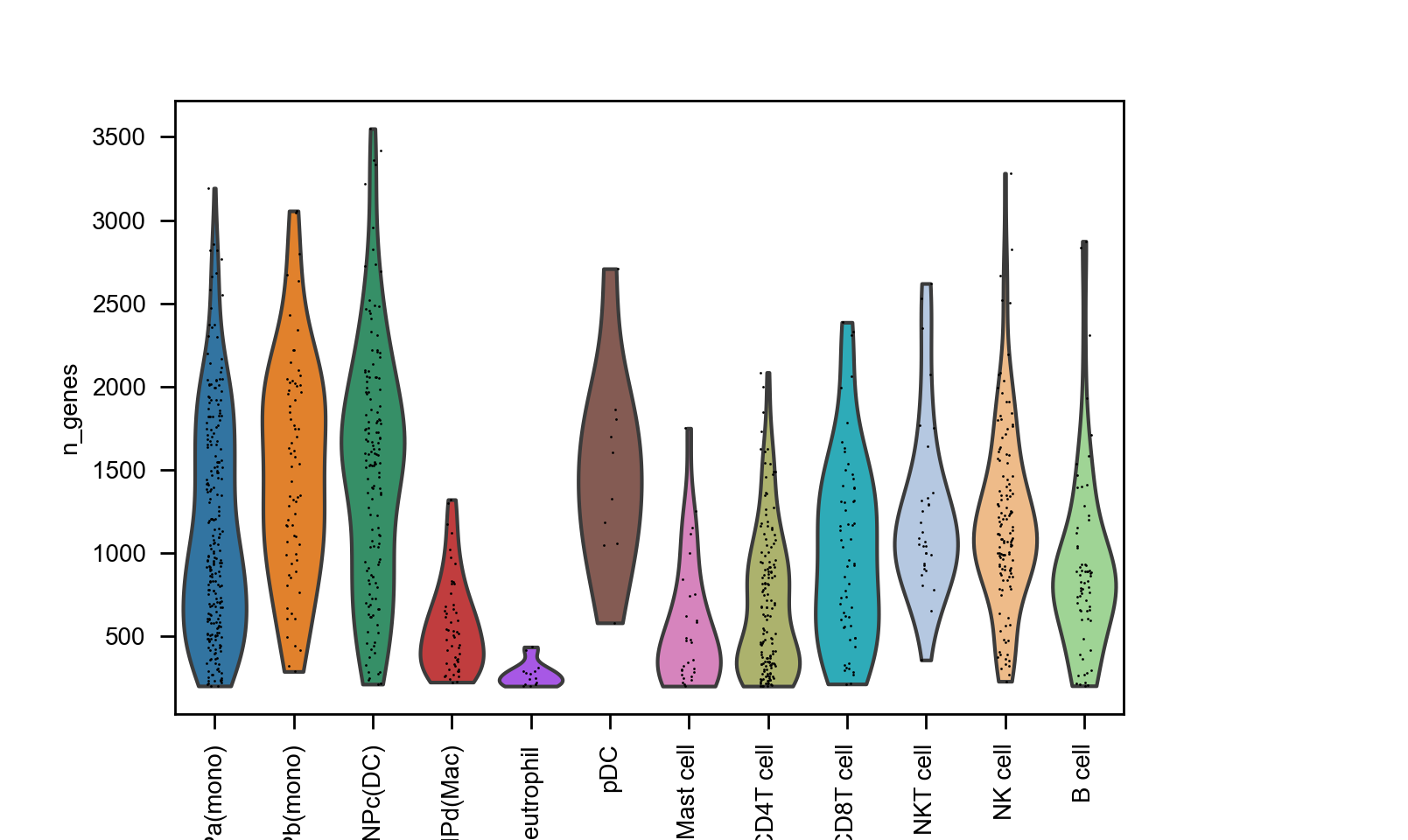
import scanpy as sc
sc.pl.violin(adata, ["n_genes"], groupby="celltype", rotation=90)
kpy.plot_cpdb_chord(
adata=adata,
cell_type1="B cell",
cell_type2=".",
means=means,
pvals=pvals,
deconvoluted=decon,
celltype_key="celltype",
genes=["PTPRC", "TNFSF13B", "BMPR2"],
figsize=(6, 6),
labelposition=50,
)
[13]:
<pycircos.pycircos.Gcircle at 0x297686890>
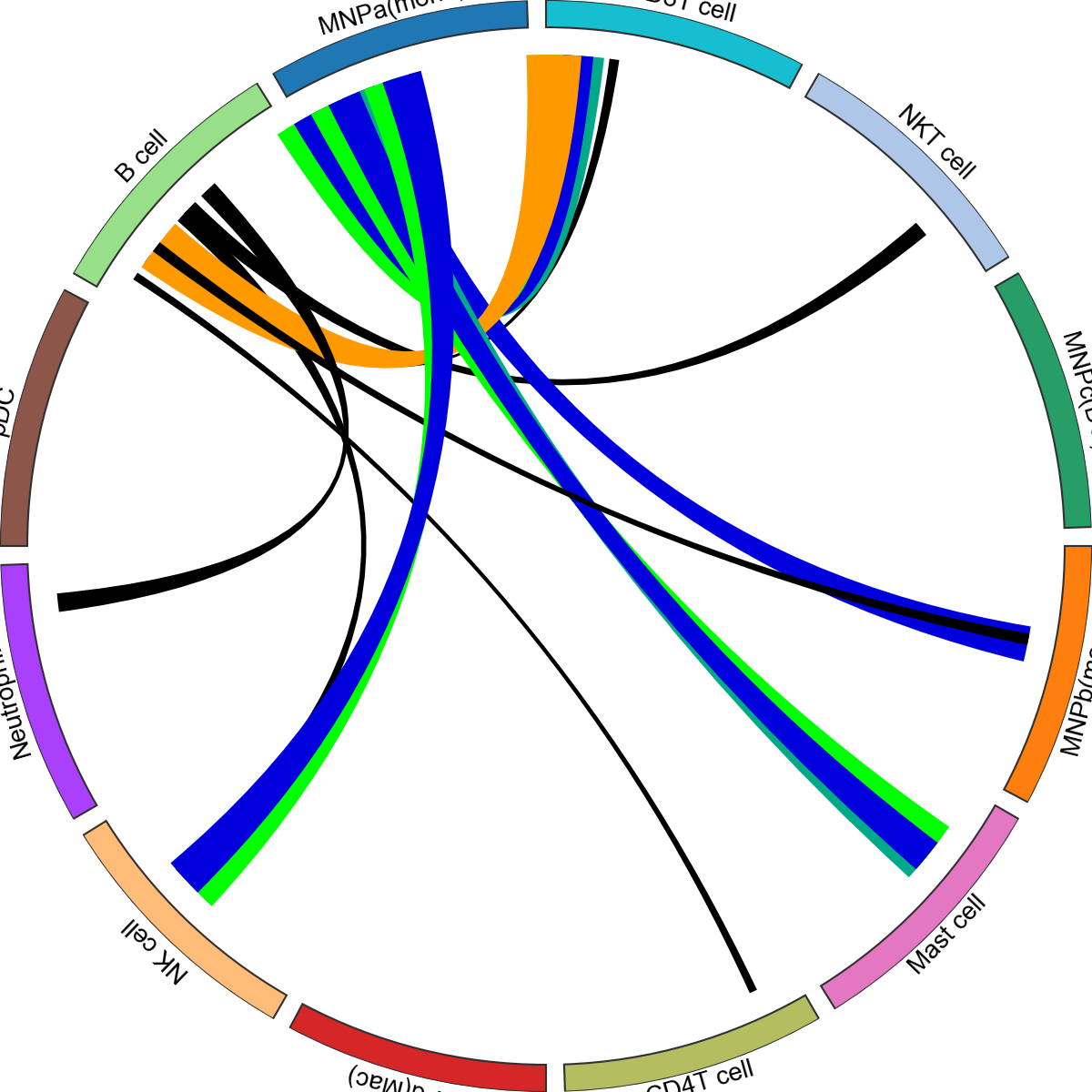
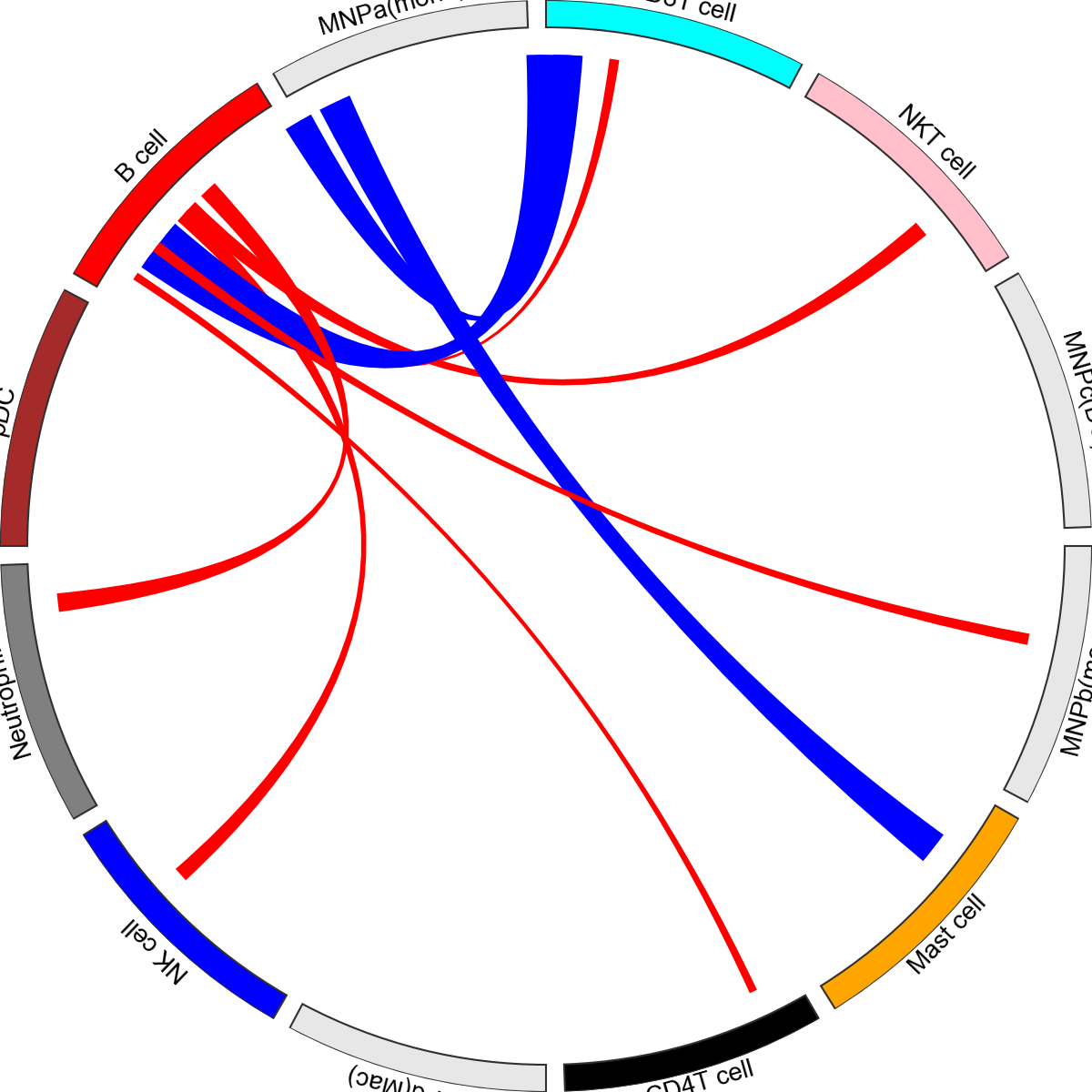
You can also provide dictionaries to change the colours for both faces and edges.
[14]:
kpy.plot_cpdb_chord(
adata=adata,
cell_type1="B cell",
cell_type2=".",
means=means,
pvals=pvals,
deconvoluted=decon,
celltype_key="celltype",
genes=["PTPRC", "TNFSF13B", "BMPR2"],
face_col_dict={
"B cell": "red",
"NK cell": "blue",
"CD4T cell": "black",
"pDC": "brown",
"Neutrophil": "grey",
"Mast cell": "orange",
"NKT cell": "pink",
"CD8T cell": "cyan",
},
edge_col_dict={"CD22-PTPRC": "red", "TNFSF13B-TNFRSF13B": "blue"},
figsize=(6, 6),
labelposition=50,
)
[14]:
<pycircos.pycircos.Gcircle at 0x297693e90>
Saving the plots
For plot_cpdb, because it’s written with plotnine, you need to save it as follows:
p = plot_cpdb(...)
p.save(...)
see also: https://plotnine.readthedocs.io/en/stable/generated/plotnine.ggplot.html
For other functions, you can use seaborn/matplotlib saving conventions e.g. plt.savefig
That’s it for now! Please check out the original ktplots R package if you are after other kinds of visualisations.
[ ]: